map run and analysis
Map run and analysis of doublet simulations¶
This is simple example of how to use the calculate_doublet_performance function from
the pythermogis package to run the simulation for a set of maps and
p-values.
This example demonstrates the following steps:
-
Reading input grids and resampling at 5km cell size for:
- Mean thickness
- Thickness standard deviation
- Net-to-gross ratio
- Porosity
- Mean permeability
- Log(permeability) standard deviation
-
Running doublet simulations and calculating economic indicators (e.g., power production, CAPEX, NPV) across all non-NaN cells in the input grids for a range of p-values (10% to 90% in 10% increments).
-
Exporting results to file.
-
Plotting result maps for selected p-values (10%, 50%, and 90%) for:
- Power output
- Capital expenditure (CAPEX)
- Net Present Value (NPV)
-
Plotting an NPV curve at a single specified location on the grid.
Example input data for this workflow is available in the /resources/example_data directory of the repository.
This example corresponds to test case test_example6 in test_doc_examples.py in the tests
directory of the repository.
from pythermogis import calculate_doublet_performance_stochastic, calculate_pos
from pygridsio import read_grid, plot_grid, resample_grid
from matplotlib import pyplot as plt
import numpy as np
import xarray as xr
from pathlib import Path
from os import path
# the location of the input data: the data can be found in the resources/example_data directory of the repo
input_data_path = Path(__file__).parent.parent / "resources" / "test_input" / "example_data"
# create a directory to write the output files to
output_data_path = Path(__file__).parent.parent / "resources" / "test_output" / "example6"
output_data_path.mkdir(parents=True, exist_ok=True)
# if set to True then simulation is always run, otherwise pre-calculated results are read (if available)
run_simulation = True
# grids can be in .nc, .asc, .zmap or .tif format: see pygridsio package
new_cellsize = 5000 # in m, this sets the resolution of the model; to speed up calcualtions you can increase the cellsize
reservoir_properties = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__thick.zmap"), new_cellsize=new_cellsize).to_dataset(name="thickness_mean")
reservoir_properties["thickness_sd"] = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__thick_sd.zmap"), new_cellsize=new_cellsize)
reservoir_properties["ntg"] = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__ntg.zmap"), new_cellsize=new_cellsize)
reservoir_properties["porosity"] = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__poro.zmap"), new_cellsize=new_cellsize) / 100
reservoir_properties["depth"] = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__top.zmap"), new_cellsize=new_cellsize)
reservoir_properties["ln_permeability_mean"] = np.log(resample_grid(read_grid(input_data_path / "ROSL_ROSLU__perm.zmap"), new_cellsize=new_cellsize))
reservoir_properties["ln_permeability_sd"] = resample_grid(read_grid(input_data_path / "ROSL_ROSLU__ln_perm_sd.zmap"), new_cellsize=new_cellsize)
# output inputs
variables_to_plot = ["depth", "thickness_mean", "thickness_sd"]
fig, axes = plt.subplots(nrows=len(variables_to_plot), ncols=1, figsize=(7, 5*len(variables_to_plot)), sharex=True, sharey=True)
for i, variable in enumerate(variables_to_plot):
plot_grid(reservoir_properties[variable], axes=axes[i], add_netherlands_shapefile=True) # See documentation on plot_grid in pygridsio, you can also provide your own shapefile
plt.tight_layout() # ensure there is enough spacing
plt.savefig(output_data_path / "input_maps_1.png")
variables_to_plot = [ "porosity", "ln_permeability_mean", "ln_permeability_sd"]
fig, axes = plt.subplots(nrows=len(variables_to_plot), ncols=1, figsize=(7, 5*len(variables_to_plot)), sharex=True, sharey=True)
for i, variable in enumerate(variables_to_plot):
plot_grid(reservoir_properties[variable], axes=axes[i], add_netherlands_shapefile=True) # See documentation on plot_grid in pygridsio, you can also provide your own shapefile
plt.tight_layout() # ensure there is enough spacing
plt.savefig(output_data_path / "input_maps_2.png")
# simulate
# if doublet simulation has already been run then read in results, or run the simulation and write results out
if (output_data_path / "output_results.nc").exists and not run_simulation:
results = xr.load_dataset(output_data_path / "output_results.nc")
else:
results = calculate_doublet_performance_stochastic(reservoir_properties, p_values=[10, 20, 30, 40, 50, 60, 70, 80, 90])
results.to_netcdf(output_data_path / "output_results.nc") # write doublet simulation results to file
# plot results as maps
variables_to_plot = ["transmissivity", "power", "pres", "npv"]
p_values_to_plot = [10, 50, 90]
fig, axes = plt.subplots(nrows=len(variables_to_plot), ncols=len(p_values_to_plot), figsize=(5 * len(variables_to_plot), 5 * len(p_values_to_plot)), sharex=True, sharey=True)
for j, p_value in enumerate(p_values_to_plot):
results_p_value = results.sel(p_value=p_value)
for i, variable in enumerate(variables_to_plot):
plot_grid(results_p_value[variable], axes=axes[i, j], add_netherlands_shapefile=True) # See documentation on plot_grid in pygridsio, you can also provide your own shapefile
plt.tight_layout() # ensure there is enough spacing
plt.savefig(output_data_path / "maps.png")
# plot net-present value at a single location as a function of p-value and find the probability of success
results["pos"] = calculate_pos(results) # calculate probability of success across whole aquifer
x, y = 125e3, 525e3 # define location
results_loc = results.sel(x=x, y=y, method="nearest") # select only the location of interest
pos = results_loc.pos.data # get probability of success at location of interest as a single value
# plot npv versus p-value and a map showing the location of interest
fig, axes = plt.subplots(ncols=2, figsize=(10, 5))
results_loc.npv.plot(y="p_value", ax=axes[0])
axes[0].set_title(f"Aquifer: ROSL_ROSLU\nlocation: [{x:.0f}m, {y:.0f}m]")
axes[0].axhline(pos, label=f"probability of success: {pos:.1f}%", ls="--", c="tab:orange")
axes[0].axvline(0.0, ls="--", c="tab:orange")
axes[0].legend()
axes[0].set_xlabel("net-present-value [Million €]")
axes[0].set_ylabel("p-value [%]")
plot_grid(results.pos, axes=axes[1], add_netherlands_shapefile=True)
axes[1].scatter(x, y, marker="x", color="tab:red")
plt.tight_layout() # ensure there is enough spacing
plt.savefig(output_data_path / "pos.png")
The figure of input maps generated by the above code will look like this:
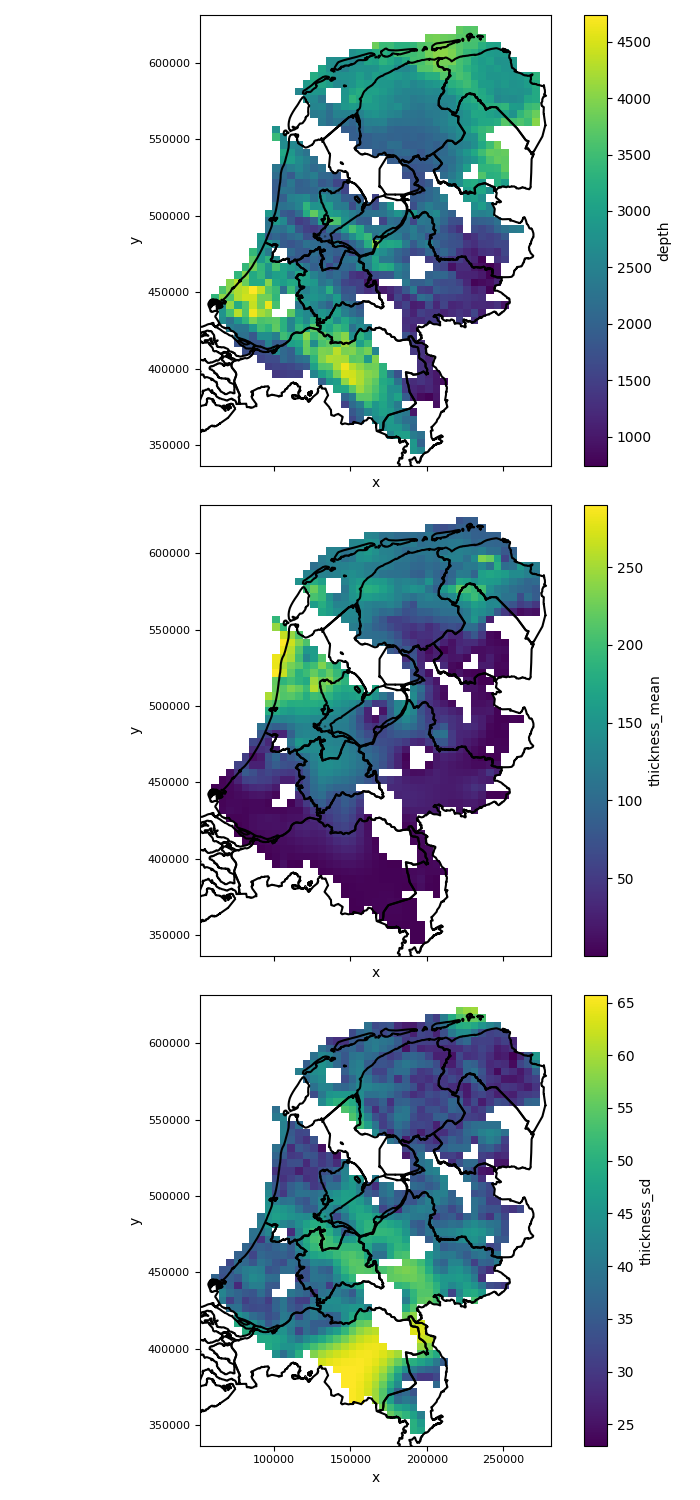 |
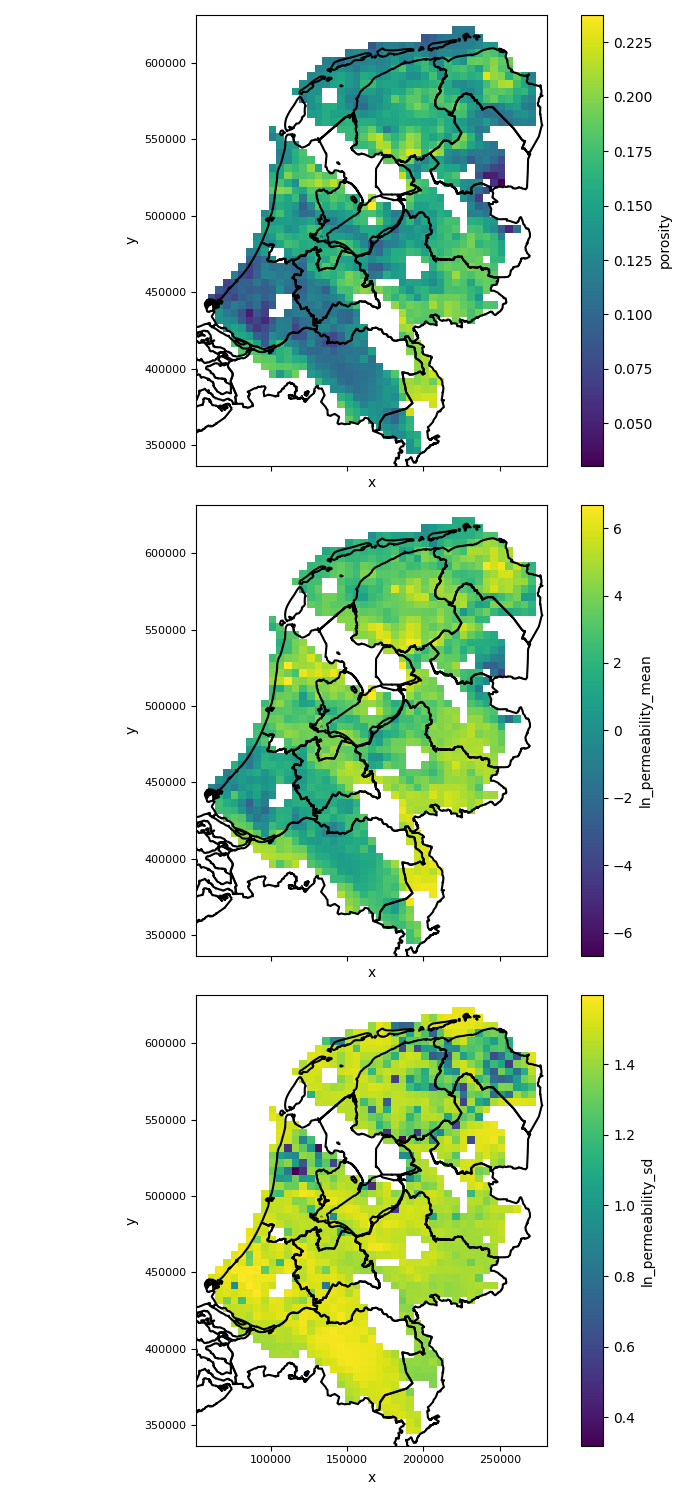 |
|---|---|
The figure generated by the above code will look like this:
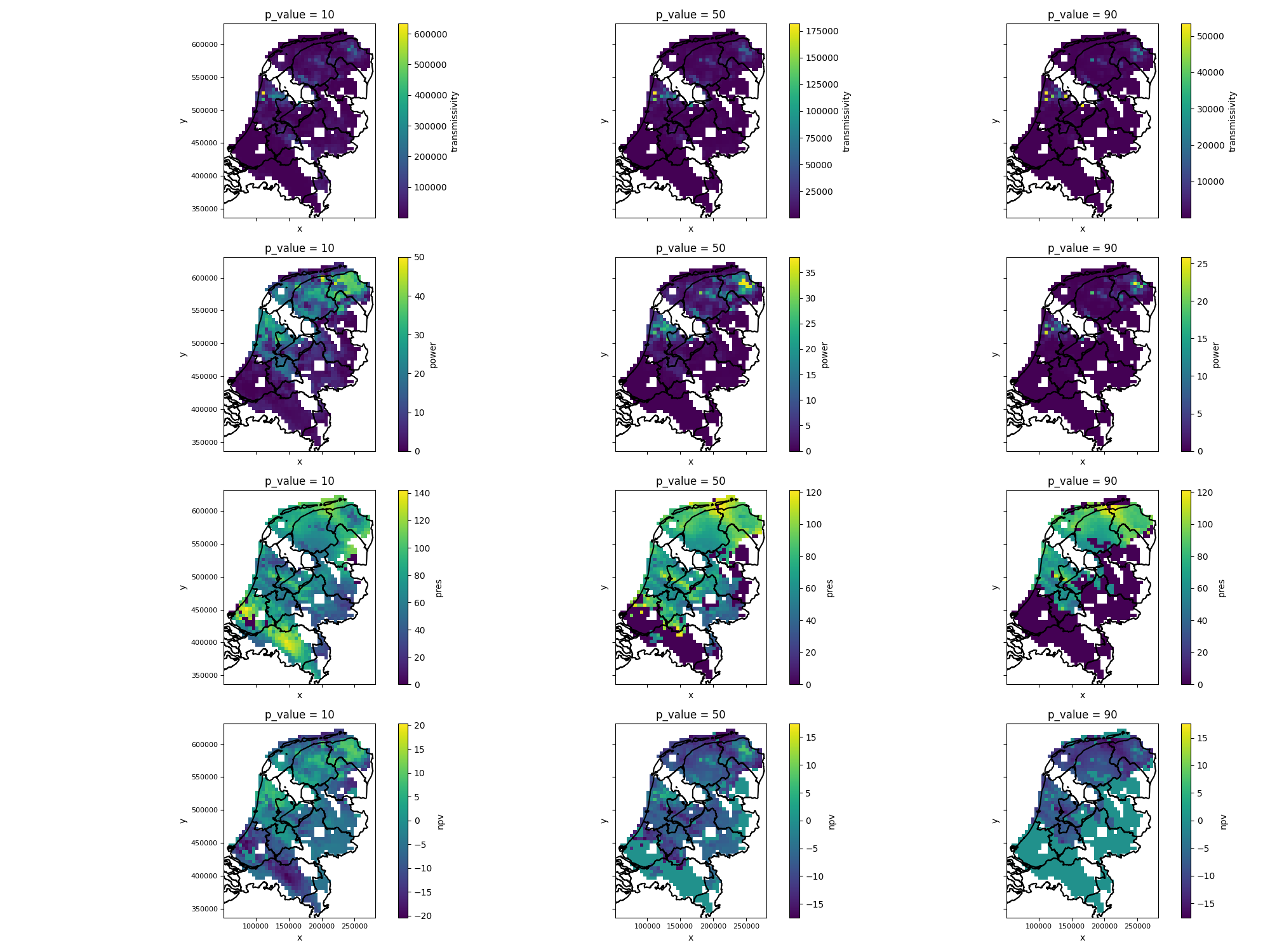
Figure: expectation maps for p-value tresholds
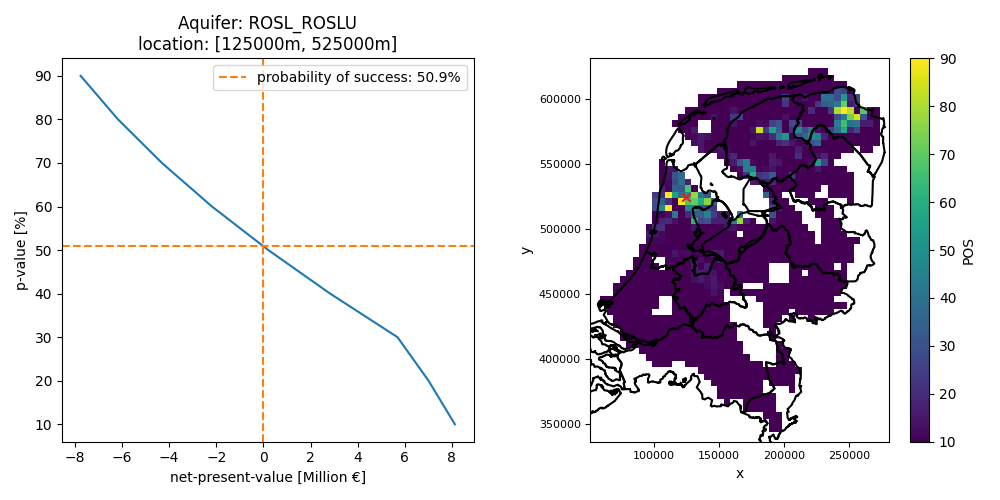 Figure: probability of success based on an NPV expectation curve at a single location
Figure: probability of success based on an NPV expectation curve at a single location